

Designing and developing targeted protein degraders is challenging for several reasons. The proteins targeted for degradation, often linked to diseases, exhibit intricate structures and functions that defy easy pinpointing of vulnerable regions or traits. Even when target sites within the protein and the E3 ligase are identified, the creation and synthesis of inhibitor ligands capable of fully activating the proteasome-dependent degradation process pose substantial challenges. The key challenge emerges during the design of the linker that connects these inhibitor ligands. Achieving the right balance in terms of composition, length, and rigidity becomes imperative in order to synthesis stable degraders with the necessary attributes of permeability and solubility.
This case study showcases a project executed by Aragen Life Sciences’ Integrated Drug Discovery (IDD) team. They successfully identified degrader molecules exhibiting the necessary physicochemical characteristics, including stability, permeability, and solubility, crucial for effectively degrading the target Protein of Interest (POI).
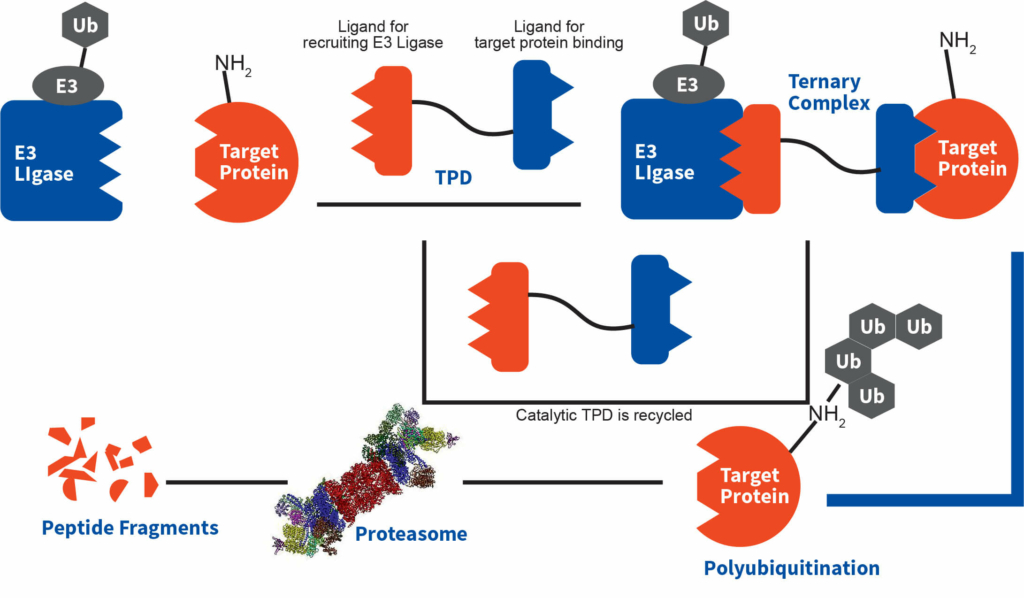
Figure. Degradation mechanism of TPDs
TPDs are heterobifunctional small molecules comprising of two active moieties (one engages E3 ubiquitin ligase and other binds to target protein (POI), meant for degradation) and a linker connecting these two active moieties involved in engaging specific unwanted protein.
The client is a precision therapy company headquartered in the Massachusetts, United States. They specialize in developing novel medications for cancer and blood disorders. Their advanced initiatives encompass systemic mastocytosis, lung cancer, and various cancers characterized by specific genomic profiles, in addition to their work in the field of cancer immunotherapy.
Project objectives were achieved through the implementation of computational and medicinal chemistry approaches. Computational approach was adopted to develop binary complexes of POI and E3-Ligase using protein-protein docking and MD (Molecular Dynamis) simulations. The optimized binary complex obtained after the MD simulation was utilized for docking with the library of enumerated linkers. Selection and prioritization of binary complexes was dependent on protein-protein interactions, alignment, and binding energy of the docked species. Preferred and non-preferred complexes obtained from the analysis were prioritized for the synthesis.
By medicinal chemistry approach, attachment sites of linkers and CRBN ligands were proposed through the understanding of structure activity relationship and alignment of linkers in the binary complex. A library of linkers consisting of linear (PEG, alkyl etc), semi-rigid and rigid types, were explored for the understanding of Stability (chemical, plasma and microsome), solubility and permeability. PHI2 (flexibility descriptor) was utilized as one of the key descriptors for linkers selection. Once the entire library was screened, attachment sites were accurately predicted, TPDs showing good physicochemical and pharmacokinetic properties were selected for the refining computational chemistry approach.

Optimization of docking protocol: extensive optimization of docking parameters was done and out of 88 experimentally designed TPDs, 77 were subjected for docking calculations. 73% TPDs were correctly predicted using the optimized parameters. Moreover, suitable length and composition of the linker for potent degrader and cellular activity was also determined. Poor solubility and permeability was attributed to thalidomide and liner linkers.
Improving stability and permeability: Modification of the linker from linear to semi-rigidification by introduction of piperazine and triazole units at various position of the linker. Cereblon binder was changed from Thalidomide to Lenalidomide and Phenyl-glutarimide (PG) to improve permeability and maintain the degradation and cellular activity.
Addressing solubility challenges: Further rigidification of the linkers by introduction of appropriate functional groups (cyclic) to increase the rigidity of the TPDs resulting in fivefold improvement in the solubility, activity, and stability (chemical, plasma and microsomal) as confirmed by various biological screening assays. Further modifications in the linkers were done to improve the solubility of the molecules at pH 6.5 enabling intestinal permeability.
The TPDs that were identified underwent rigorous potency testing, demonstrating remarkable effectiveness, as evidenced by the in vitro assays that are used to quantify the Protein of Interest (POI). The parameters optimized in the computational approach proved to be nearly five times more efficient than existing methods for generating TPD conformers. The project was not only successfully delivered to the client but also received approval for further development. This next phase involves subjecting the identified TPDs to pre-clinical trials, marking a significant advancement in the project’s progression.
