

In drug development, distribution coefficient (LogD) is an important attribute that describes the lipophilicity of a drug candidate. It is a physicochemical characteristic of a compound that influences its ability to permeate biological membranes, thereby affecting its absorption, distribution, metabolism, and excretion (ADME) properties. Since it reflects both ionized and unionized forms, LogD provides a more accurate reflection of a drug’s behavior in biological systems. The gold-standard method for determining LogD is the traditional shake flask method, however, being slower and more labor-intensive, it is cumbersome for a high-throughput format. Furthermore, although LogD can be predicted via several in silico models, there are several challenges (Figure 1) for performing these.
Figure 1: Challenges in predicting LogD.
Thus, to rapidly generate high-quality LogD data to support in silico predictions during drug discovery process, a high-throughput technique like high-performance liquid chromatography and tandem mass spectrometry (LC-MS/MS) is the method of choice.
Aragen’s scientists established a high-throughput screening assay for LogD determination that combines the traditional shake flask method with LC-MS/MS analysis. We simultaneously analyzed 11 quality control (QC) compounds of diverse chemistry. Two different sample pooling approaches-post-experiment and pre-experiment were employed prior to the LC-MS/MS analysis. In the post-experiment method, each QC compound was individually subjected to the shake-flask method to separate into octanol and aqueous phases. The octanol and aqueous fractions of each QC compound were then pooled separately into different samples. This resulted in multiple samples (one for octanol phase and one for aqueous phase) for each QC compound. In the pre-experiment technique, all 11 QC compounds were mixed to form a single mixture. This mixture was then separated into octanol and aqueous phases by shake-flask method, resulting in octanol and aqueous fractions from the pooled mixture. Subsequently, the analytes from both the set of experiments were analyzed by LC-MS/MS. The method was evaluated by comparing LogD values of analytes generated by the post-experiment and pre-experiment approaches.
In the current study, our team validated a LogD assay that combines shake-flask method with LC-MS/MS for 11 QC compounds, belonging to varied lipophilicity and ionization classes. The method is useful in determining the LogD values in a high-throughput format. We used different sample pooling approaches (post-experiment versus pre-experiment) to effectively screen LogD values of multiple QC compounds using the shake-flask method, followed by a robust LC-MS/MS analysis for accurate LogD measurements. The post-experiment approach enabled individual characterization of LogD values for each QC compound, offering specific distribution data in octanol/water for detailed comparison across compounds. The LogD values obtained by post-experiment technique aligned with the previously reported literature values (Table 1).
Table 1: LogD data from the experiments (post-experiment and pre experiment) versus literature values.
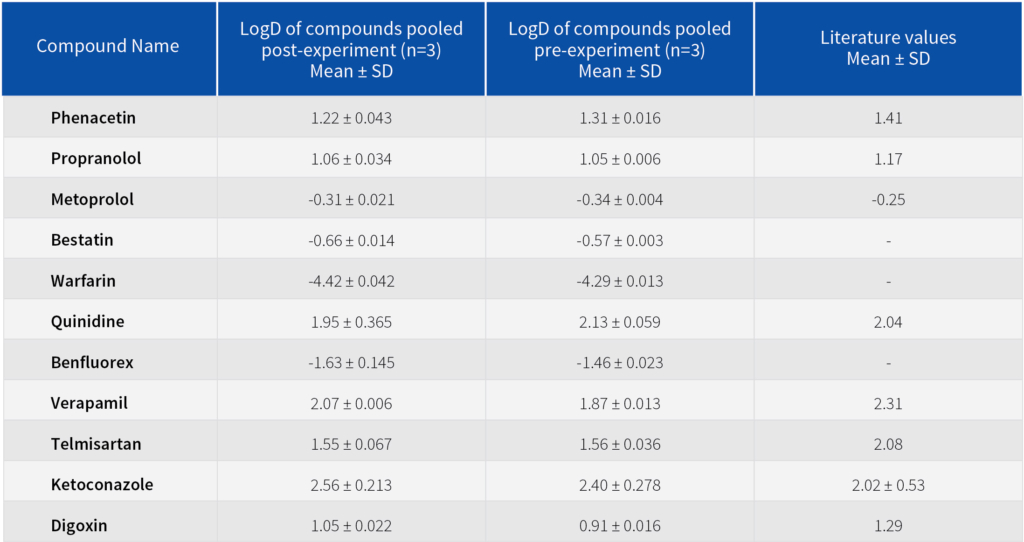
The pre-experiment technique, which pooled the 11 QC compounds before the shake-flask assay, streamlined sample processing before LC-MS/MS analysis. This pooling approach expands the effective LogD measurement range (-0.66 to 2.40) and yields reproducible data across a wide range of values (Figure 2). It also saves ~20% time compared to the post-experiment pooling technique. Additionally, the pre-experiment technique simulates a biological sample where multiple compounds are present together, revealing collective distribution in octanol/water, ideal for screening overall behavior rather than individual compounds. The LogD values obtained by pre-experiment technique are given in the table 1 and are comparable with the post-experiment technique LogD values.
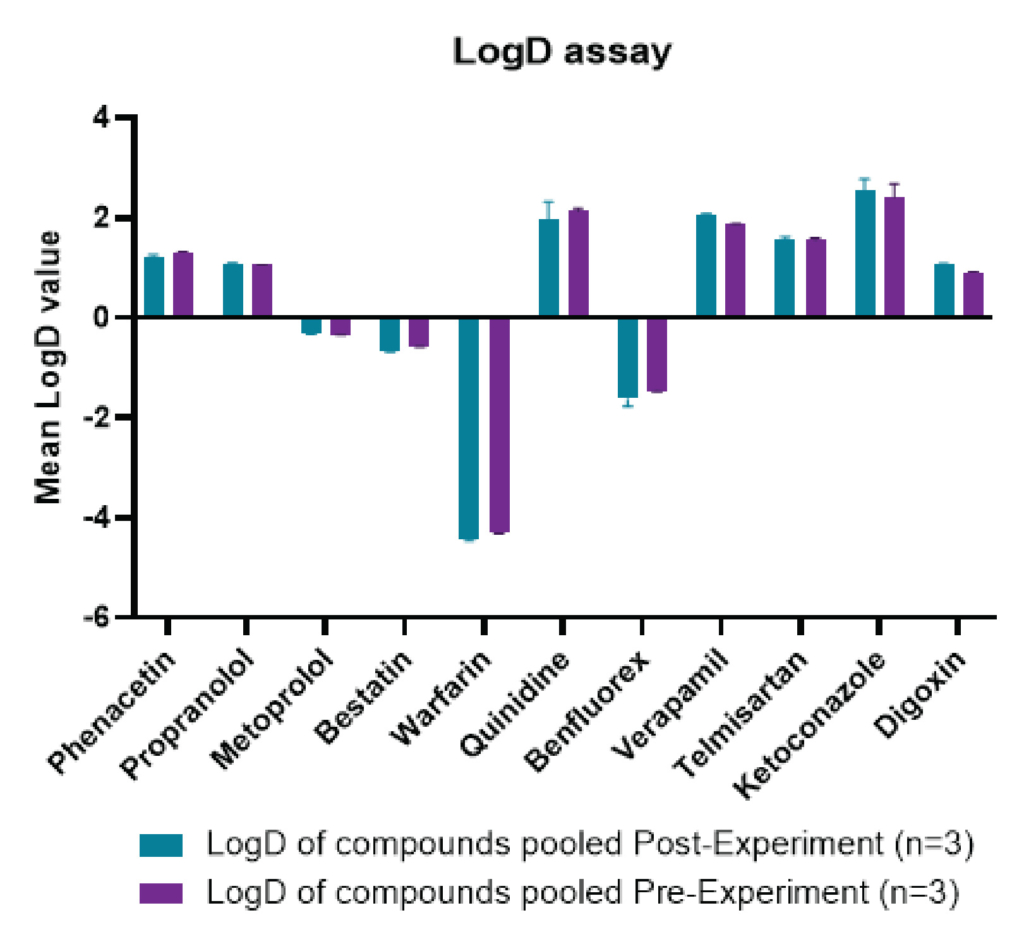
Figure 2: A graph comparing the LogD values of the pre-experiment and in post-experiment analytes. Each test drug was analyzed in triplicates and data represents mean, SD and % CV for each set of triplicates.
Conclusively, combining the traditional shake flask method with LC-MS/MS bioanalysis and using a sample pooling approach, allowed for efficient, high-throughput screening of LogD during the drug discovery process. High-throughput beside allowing rapid testing of multiple compounds simultaneously generates consistent and reproducible data and are cost-effective. Furthermore, in silico LogD predictions can be refined by comparing them with the real-world experimental data, facilitating more accurate and relevant predictions for guiding drug discovery and development processes.
Aragen’s biologists are equipped to address the varied drug discovery requirements of our global life sciences clientele and with our team’s expertise we offer:
If you’re interested in DMPK excellence, Aragen is ready to accelerate your drug discovery journey!

